Developing CODE for a Research Database
The fact that there are a plethora of scientific papers readily available online would seem helpful to researchers. Unfortunately, the truth is that the volume of these articles has grown such that determining which information is relevant to a specific project is becoming increasingly difficult.
 Austrian and German researchers are thus developing CODE, or Commercially Empowered Linked Open Data Ecosystems in Research, to properly aggregate research data from its various forms, such as PDFs of academic papers and data tables upon which those papers are based, into a single system. The project is in a prototype stage, with the goal being to integrate all forms into one platform by the project’s second year.
Austrian and German researchers are thus developing CODE, or Commercially Empowered Linked Open Data Ecosystems in Research, to properly aggregate research data from its various forms, such as PDFs of academic papers and data tables upon which those papers are based, into a single system. The project is in a prototype stage, with the goal being to integrate all forms into one platform by the project’s second year.
The researchers from the University of Passau in Germany and the Know-Center in Graz, Austria explored the challenges to CODE and how the team intends to deal with those challenges in this paper. The goal is to meliorate the research process by making it easier to not only search for both text and numerical data in the same query but also to use both varieties in concert. The basic architecture for the project is shown below.
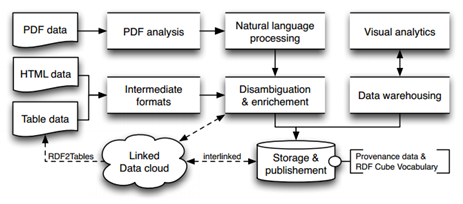
The cumbersome nature of the task at hand is apparent by scanning the left side of the diagram. The PDF data refers to the actual text from scientific research papers that needs to be analyzed semantically. The HTML data refers to the metadata located within the document while the Table data references the numerical and statistical data behind the paper. An oversimplified view sees that, for the most part, the PDF text data is unstructured, the table data is structured, and the metadata is some combination of the two.
“As already shown, in the CODE project a high number of diverse as well as highly dynamic data sources are present, each offering huge portions of data,” as described in the paper. “Obviously, this situation leads to (an) integration problem with uncertainty in terms of data quality.” CODE’s goal will be to isolate each approach, individually prepare them, and then integrate them into a master strategy.
A good example of where CODE is crowd-sourcing a problem is PDF analysis, which the paper noted can be tricky. For one, PDF text data is organized in a manner that does not necessarily follow human logic. One can notice a propagation of this if they try copying text from a PDF onto a word document; the lines break at odd places. While this can easily be fixed by an attentive human user and this exact problem is unlikely to harbor itself in CODE’s research, it represents incongruence between formatting styles that have to be dealt with over and over across a range of scientific papers.
As mentioned above, CODE sees crowd-sourcing as the tool for providing a workable solution, especially given the different methods already discovered for particular aspects of PDF text integration. “Despite the progress in the single steps, there is no general solution that can provide all information in the quality needed within the CODE project in sufficient quality. Thus, the task is to aggregate results from recent research on PDF analysis into the CODE prototype and adapt or refine existing approaches.”
It should be noted that while CODE’s work is essentially a big data problem, the resulting platform will not exist solely for big data research. This concept could actually bridge the various disciplines as CODE takes research that works in both computer science and genomics, for example, and combines them.
The paper explains how computer science research would be annotated and earmarked in the overall CODE system: “In the computer science domain, where ontologies and annotated corpora are scarce, the users of the CODE platform themselves annotate the relevant concepts. Starting with the automatic detection of named entities, the relationship between those are identified in a second step.”
The step of actually combining the data into the structure shown above is challenging. As such, it is this portion of the project where the most works seems to be needed. For example, the paper mentions the integration of RDF cubes, but notes that current research falls short with regard to interconnection. According to CODE, “high-level analytical retrieval in graph structures” will also be an essential, and somewhat unknown, aspect of the final system.
The interconnects seem to be the largest stumbling block in this ambitious project, as CODE has already found a storage framework (in Linked Media Framework) while specific modeling efforts are underway and ready to be implemented. “For data modeling tasks, two W3C standardization efforts are in scope, which will be soon issued as official recommendations…Further, the W3C proposes the RDF Cube Vocabulary as foundation for data cubes, which are the foundation of data warehouses.”
Once the data is interconnected, CODE will hope to achieve easy access to information via what they call a Vizualization Wizard. The Wizard, which will be powered by MindMeister from Meister Labs, would do what general visualization software can accomplish today: generating charts and graphs where the program helps the user find correlation.
A lot of work has to be done to integrate the wealth of scientific data on the internet that CODE is planning on aggregating. This is not news, however, as the paper’s intention was indeed to inform the work that is to be done over the next couple of years. If CODE can successfully integrate all of the various scientific data into one system, the effort will be worthwhile.
Related Articles